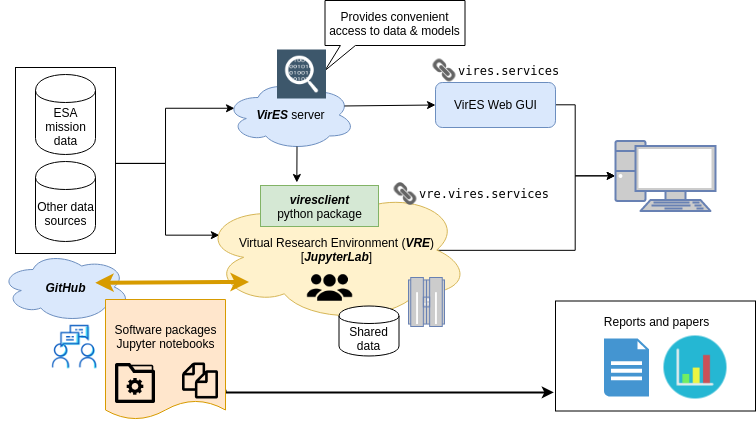
viresclient: Programmatic access to Swarm¶
ashley.smith@ed.ac.uk ... View slides: https://bit.ly/2lYAuQm
Aims¶
Direct access to "computation-ready" data without worrying about:
- file formats; file organisation
- model forward code
Provide a dependable interface to a wide arrange of "data"
- new products added / old products changed: access them in the same way
Complementary to the VirES web interface
- More prerequisite knowledge needed (++time)
- ... but more freedom than the GUI
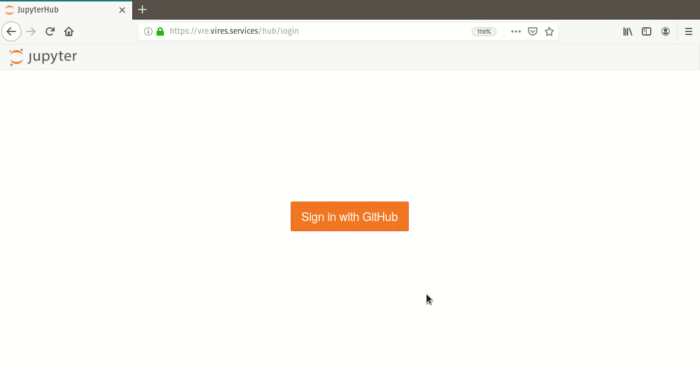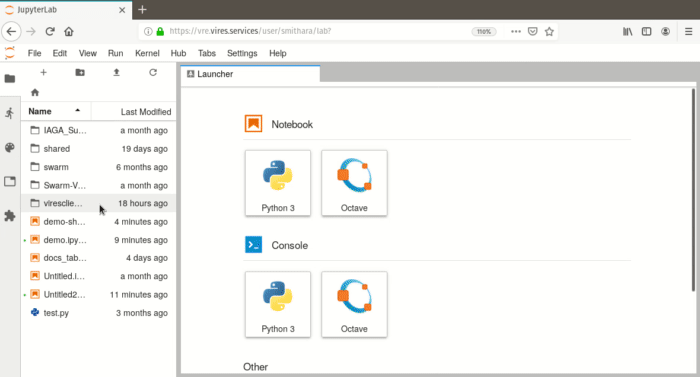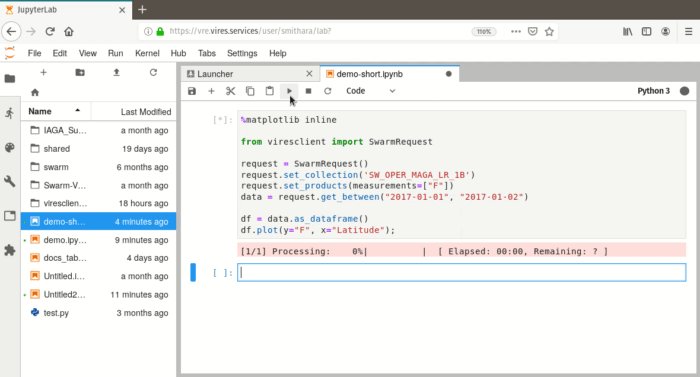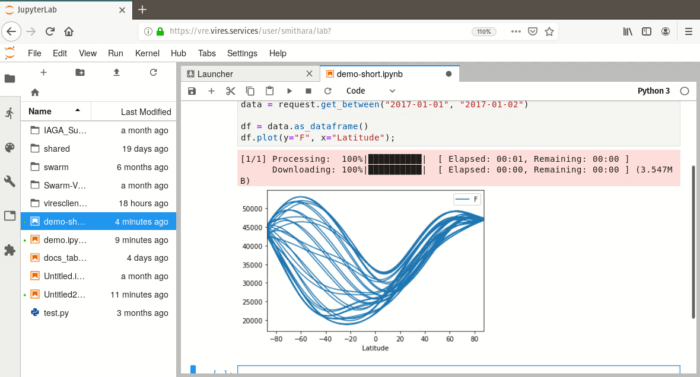
1: Basic usage: accessing data¶
In [2]:
from viresclient import SwarmRequest
request = SwarmRequest()
request.set_collection("SW_OPER_MAGA_LR_1B")
request.set_products(["F", "B_NEC"])
data = request.get_between("2019-01-01", "2019-01-02")
print(data)
[1/1] Processing: 100%|██████████| [ Elapsed: 00:01, Remaining: 00:00 ]
Downloading: 100%|██████████| [ Elapsed: 00:00, Remaining: 00:00 ] (5.621MB)
viresclient ReturnedData object of type cdf
Save it to a file with .to_file('filename')
Load it as a pandas dataframe with .as_dataframe()
Load it as an xarray dataset with .as_xarray()
In [3]:
data.to_file("test_file.cdf", overwrite=True)
Data written to test_file.cdf
2: Basic usage: translating to a Pandas dataframe¶
In [4]:
df = data.as_dataframe(expand=True)
df.head()
Out[4]:
| Spacecraft | Latitude | Longitude | Radius | F | B_NEC_N | B_NEC_E | B_NEC_C | |
|---|---|---|---|---|---|---|---|---|
| 2019-01-01 00:00:00 | A | -17.029902 | -136.020760 | 6819106.81 | 27177.2411 | 23675.8346 | 5295.5528 | -12248.4860 |
| 2019-01-01 00:00:01 | A | -16.965741 | -136.021687 | 6819098.76 | 27160.4330 | 23685.5394 | 5290.1805 | -12194.6394 |
| 2019-01-01 00:00:02 | A | -16.901579 | -136.022616 | 6819090.71 | 27143.6815 | 23695.2730 | 5284.5827 | -12140.7419 |
| 2019-01-01 00:00:03 | A | -16.837417 | -136.023547 | 6819082.65 | 27127.0045 | 23705.0150 | 5278.8028 | -12086.7943 |
| 2019-01-01 00:00:04 | A | -16.773255 | -136.024480 | 6819074.58 | 27110.3737 | 23714.6933 | 5273.2987 | -12032.8263 |
In [5]:
%matplotlib inline
df["F"].plot();
Some very short Pandas examples....¶
In [6]:
df.describe()
Out[6]:
| Latitude | Longitude | Radius | F | B_NEC_N | B_NEC_E | B_NEC_C | |
|---|---|---|---|---|---|---|---|
| count | 86400.000000 | 86400.000000 | 8.640000e+04 | 86400.000000 | 86400.000000 | 86400.000000 | 86400.000000 |
| mean | 1.155294 | -1.629096 | 6.815232e+06 | 37410.442724 | 14772.935307 | 60.615997 | 1848.167209 |
| std | 51.787736 | 104.273714 | 6.615887e+03 | 9144.052183 | 9190.478710 | 4844.955323 | 33964.585152 |
| min | -87.346563 | -179.995322 | 6.804485e+06 | 18823.264000 | -11667.202400 | -12913.468900 | -53194.233500 |
| 25% | -43.785362 | -87.030647 | 6.808636e+06 | 29574.217075 | 8659.573700 | -2833.234175 | -29038.685625 |
| 50% | 1.760706 | -7.505913 | 6.816694e+06 | 38669.085350 | 14962.491550 | 137.742450 | -986.058400 |
| 75% | 46.453081 | 87.254043 | 6.821806e+06 | 45895.150975 | 22069.236525 | 3170.253025 | 37875.310025 |
| max | 87.346203 | 179.994337 | 6.823164e+06 | 53266.250300 | 32982.012300 | 12372.612300 | 49093.207000 |
In [7]:
from pandas.plotting import autocorrelation_plot
df["F"].resample("60s").mean().pipe(autocorrelation_plot);
You can still directly access Numpy arrays from a dataframe:
In [8]:
df[["B_NEC_N", "B_NEC_E", "B_NEC_C"]].values
Out[8]:
array([[ 23675.8346, 5295.5528, -12248.486 ],
[ 23685.5394, 5290.1805, -12194.6394],
[ 23695.273 , 5284.5827, -12140.7419],
...,
[ 18781.8569, 2075.3522, 36066.841 ],
[ 18811.1275, 2073.3059, 36030.6969],
[ 18840.3446, 2071.3268, 35994.451 ]])
3. Basic usage: translate to an xarray Dataset¶

In [9]:
ds = data.as_xarray()
ds
Out[9]:
<xarray.Dataset>
Dimensions: (B_NEC_dim1: 3, Timestamp: 86400)
Coordinates:
* Timestamp (Timestamp) datetime64[ns] 2019-01-01 ... 2019-01-01T23:59:59
Dimensions without coordinates: B_NEC_dim1
Data variables:
Spacecraft (Timestamp) <U1 'A' 'A' 'A' 'A' 'A' 'A' ... 'A' 'A' 'A' 'A' 'A'
Latitude (Timestamp) float64 -17.03 -16.97 -16.9 ... 44.06 44.0 43.93
Longitude (Timestamp) float64 -136.0 -136.0 -136.0 ... 40.88 40.88 40.88
Radius (Timestamp) float64 6.819e+06 6.819e+06 ... 6.809e+06 6.809e+06
F (Timestamp) float64 2.718e+04 2.716e+04 ... 4.07e+04 4.068e+04
B_NEC (Timestamp, B_NEC_dim1) float64 2.368e+04 ... 3.599e+04
Attributes:
Sources: ['SW_OPER_MAGA_LR_1B_20190101T000000_20190101T235959_050...
MagneticModels: []
RangeFilters: []
In [10]:
ds["B_NEC"].plot.line(figsize=(10,5), x="Timestamp");
4. Robust and easy access to larger volumes of data and models¶
In [11]:
request = SwarmRequest()
request.set_collection("SW_OPER_MAGA_LR_1B")
request.set_products(
measurements=["B_NEC"], models=["CHAOS-6-Core"], residuals=True,
auxiliaries=["MLT", "QDLat"], sampling_step="PT60S")
request.set_range_filter("Flags_F", 0, 1)
data = request.get_between("2019-01-01", "2019-07-01") # 6 MONTHS
df = data.as_dataframe(expand=True)
df.plot(y="B_NEC_res_CHAOS-6-Core_C", x="QDLat", kind="scatter", figsize=(10,3),
c="MLT", cmap=cm.RdYlBu, s=1, alpha=0.5);
[1/1] Processing: 100%|██████████| [ Elapsed: 00:18, Remaining: 00:00 ]
Downloading: 100%|██████████| [ Elapsed: 00:00, Remaining: 00:00 ] (17.746MB)
Names of original data files are logged¶
In [12]:
data.sources[-10:]
Out[12]:
['SW_OPER_MAGA_LR_1B_20190623T000000_20190623T235959_0505_MDR_MAG_LR', 'SW_OPER_MAGA_LR_1B_20190624T000000_20190624T235959_0505_MDR_MAG_LR', 'SW_OPER_MAGA_LR_1B_20190625T000000_20190625T235959_0505_MDR_MAG_LR', 'SW_OPER_MAGA_LR_1B_20190626T000000_20190626T235959_0505_MDR_MAG_LR', 'SW_OPER_MAGA_LR_1B_20190627T000000_20190627T235959_0505_MDR_MAG_LR', 'SW_OPER_MAGA_LR_1B_20190628T000000_20190628T235959_0505_MDR_MAG_LR', 'SW_OPER_MAGA_LR_1B_20190629T000000_20190629T235959_0505_MDR_MAG_LR', 'SW_OPER_MAGA_LR_1B_20190630T000000_20190630T235959_0505_MDR_MAG_LR', 'SW_OPER_MAGA_LR_1B_20190701T000000_20190701T235959_0505_MDR_MAG_LR', 'SW_OPER_MCO_SHA_2X_19970101T000000_20190911T235959_0609']
"One-line" analysis¶
In [13]:
ds = data.as_xarray()
fig, ax = plt.subplots(1, 1, figsize=(10, 2))
(ds.groupby_bins("QDLat", 90)
.apply(lambda x: x["B_NEC_res_CHAOS-6-Core"].std(axis=0))
.plot.line(x="QDLat_bins", ax=ax)
)
ax.set_title("Standard deviations");
Want to do this? Learn Pandas & Xarray (& Numpy & Matplotlib)
Want to scale it to larger data? Learn Dask.
In [15]:
from viresclient import SwarmRequest
import matplotlib.pyplot as plt
from matplotlib.dates import DateFormatter
request = SwarmRequest()
request.set_collection("SW_OPER_IPDAIRR_2F")
request.set_products(measurements=request.available_measurements("IPD"))
data = request.get_between("2014-12-21T00:00", "2014-12-21T03:00")
df = data.as_dataframe()
fig, axes = plt.subplots(nrows=7, ncols=1, figsize=(20,11), sharex=True)
df.plot(ax=axes[0], y=['Background_Ne', 'Foreground_Ne', 'Ne'], alpha=0.8)
df.plot(ax=axes[1], y=['Grad_Ne_at_100km', 'Grad_Ne_at_50km', 'Grad_Ne_at_20km'])
df.plot(ax=axes[2], y=['RODI10s', 'RODI20s'])
df.plot(ax=axes[3], y=['ROD'])
df.plot(ax=axes[4], y=['mROT'])
df.plot(ax=axes[5], y=['delta_Ne10s', 'delta_Ne20s', 'delta_Ne40s'])
df.plot(ax=axes[6], y=['mROTI20s', 'mROTI10s'])
for ax in axes:
ax.xaxis.set_major_formatter(DateFormatter("%Y-%m-%d\n%H:%M:%S"))
ax.legend(loc="upper right")
ax.grid()
fig.subplots_adjust(hspace=0)
plt.close()
[1/1] Processing: 100%|██████████| [ Elapsed: 00:01, Remaining: 00:00 ]
Downloading: 100%|██████████| [ Elapsed: 00:00, Remaining: 00:00 ] (2.273MB)
Replace the previous with:
from viresclient import SwarmQuicklook
fig = SwarmQuicklook("IPDxIRR", spacecraft="Alpha", options...)
In [16]:
fig
Out[16]:
6. Future development: integrate with other libraries¶
 |
 |
 |
 |
| PyViz advanced visualisation: |  |
| Domain-specific libraries: |  |
 |
 |
? Swarm-DISC... |
In [17]:
import hvplot.pandas
df.hvplot(y=['Background_Ne', 'Foreground_Ne', 'Ne'])
Out[17]:
7. Develop other packages which connect to viresclient¶
SwarmPyFAC (author: Ask Neve Gamby)¶
New Swarm-DISC GitHub organisation
https://github.com/Swarm-DISC/SwarmPyFAC
In [19]:
import swarmpyfac as fc
import datetime as dt
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
output, input_data = fc.fac_from_file(start=dt.datetime(2016, 1, 1), end=dt.datetime(2016, 1, 2), user_file=None)
time, position, __, fac, *___ = output
selection = np.arange(380,3000)
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(15,5))
axes[0].plot(time,position[:,0],'b')
axes[0].plot(time[selection],position[selection,0],'r')
axes[0].set_xlabel('time [s]]'); axes[0].set_ylabel('latitude [degree]')
axes[1].plot(position[selection,0],fac[selection],'b')
axes[1].set_xlabel('latitude [degree]'); axes[1].set_ylabel('$J_{||} \; [nA/m^2]$')
axes[1].axis([-90, 90, -15,15]);
8. Compatibility and versioning¶
- viresclient defines a general purpose data access layer for other packages to depend on
- Semantic versioning. Read this if you are producing a package: https://semver.org
In [20]:
import viresclient
viresclient.__version__
Out[20]:
'0.4.1'
- Forwards compatibility is a priority but can't be guaranteed right now - check the change log
- Aim to formalise interface and move to a 1.0 release when appropriate
Summary¶
- viresclient: https://viresclient.readthedocs.io/en/latest/installation.html
- VRE: https://swarm-vre.readthedocs.io
- Ongoing development of viresclient tying the VirES/VRE services together
- Aim to integrate with other Python packages
- General Python software (many 1000's of contributors)
- Swarm software from the community: Coordinate using GitHub (see e.g. sunpy)
- Need feedback and feature requests for real use cases
- Need community-led development of packages
- Use cases:
- from rapid development, to producing repeatable & portable analyses
- data access layer for other affiliated packages (Swarm-DISC activities)
- integrating VirES with other services (e.g. Swarm-Aurora/AuroraX)